Deterministic tractography
Overview
Teaching: 30 min
Exercises: 0 minQuestions
What computations does a deterministic tractography require?
How can we visualize the streamlines generated by a tractography method?
Objectives
Be able to perform deterministic tracking on diffusion MRI data
Familiarize with the data entities of a tractogram
Deterministic tractography
Deterministic tractography algorithms perform tracking of streamlines by following a predictable path, such as following the primary diffusion direction.
In order to demonstrate how to perform deterministic tracking on a diffusion MRI dataset, we will build from the preprocessing presented in a previous episode and compute the diffusion tensor.
import os
import nibabel as nib
import numpy as np
from bids.layout import BIDSLayout
from dipy.io.gradients import read_bvals_bvecs
from dipy.core.gradients import gradient_table
dwi_layout = BIDSLayout("../../data/ds000221/derivatives/uncorrected_topup_eddy", validate=False)
gradient_layout = BIDSLayout("../../data/ds000221/", validate=False)
subj = '010006'
dwi_fname = dwi_layout.get(subject=subj, suffix='dwi', extension='.nii.gz', return_type='file')[0]
bvec_fname = dwi_layout.get(subject=subj, extension='.eddy_rotated_bvecs', return_type='file')[0]
bval_fname = gradient_layout.get(subject=subj, suffix='dwi', extension='.bval', return_type='file')[0]
dwi_img = nib.load(dwi_fname)
affine = dwi_img.affine
bvals, bvecs = read_bvals_bvecs(bval_fname, bvec_fname)
gtab = gradient_table(bvals, bvecs)
We will now create a mask and constrain the fitting within the mask.
Tractography run times
Note that many steps in the streamline propagation procedure are computationally intensive, and thus may take a while to complete.
import dipy.reconst.dti as dti
from dipy.segment.mask import median_otsu
dwi_data = dwi_img.get_fdata()
dwi_data, dwi_mask = median_otsu(dwi_data, vol_idx=[0], numpass=1) # Specify the volume index to the b0 volumes
dti_model = dti.TensorModel(gtab)
dti_fit = dti_model.fit(dwi_data, mask=dwi_mask) # This step may take a while
We will perform tracking using a deterministic algorithm on tensor fields via
EuDX (Garyfallidis et al., 2012).
EuDX makes use of the primary direction of the diffusion tensor to propagate
streamlines from voxel to voxel and a stopping criteria from the fractional
anisotropy (FA).
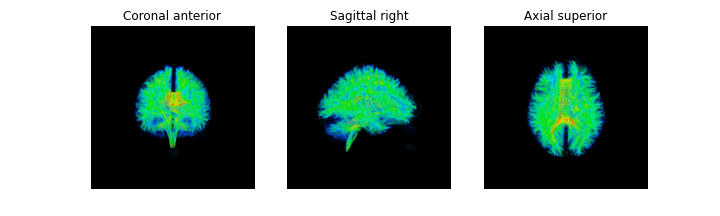
We will first get the FA map and eigenvectors from our tensor fitting. In the
background of the FA map, the fitting may not be accurate as all of the
measured signal is primarily noise and it is possible that values of NaNs (not
a number) may be found in the FA map. We can remove these using numpy to find
and set these voxels to 0.
# Create the directory to save the results
out_dir = f"../../data/ds000221/derivatives/dwi/tractography/sub-{subj}/ses-01/dwi/"
if not os.path.exists(out_dir):
os.makedirs(out_dir)
fa_img = dti_fit.fa
evecs_img = dti_fit.evecs
fa_img[np.isnan(fa_img)] = 0
# Save the FA
fa_nii = nib.Nifti1Image(fa_img.astype(np.float32), affine)
nib.save(fa_nii, os.path.join(out_dir, 'fa.nii.gz'))
# Plot the FA
import matplotlib.pyplot as plt
from scipy import ndimage # To rotate image for visualization purposes
%matplotlib inline
fig, ax = plt.subplots(1, 3, figsize=(10, 10))
ax[0].imshow(ndimage.rotate(fa_img[:, fa_img.shape[1]//2, :], 90, reshape=False))
ax[1].imshow(ndimage.rotate(fa_img[fa_img.shape[0]//2, :, :], 90, reshape=False))
ax[2].imshow(ndimage.rotate(fa_img[:, :, fa_img.shape[-1]//2], 90, reshape=False))
fig.savefig(os.path.join(out_dir, "fa.png"), dpi=300, bbox_inches="tight")
plt.show()
One of the inputs of EuDX is the discretized voxel directions on a unit
sphere. Therefore, it is necessary to discretize the eigenvectors before
providing them to EuDX. We will use an evenly distributed sphere of 362
points using the get_sphere function.
from dipy.data import get_sphere
sphere = get_sphere('symmetric362')
We will determine the indices representing the discretized directions of the peaks by providing as input, our tensor model, the diffusion data, the sphere, and a mask to apply the processing to. Additionally, we will set the minimum angle between directions, the maximum number of peaks to return (1 for the tensor model), and the relative peak threshold (returning peaks greater than this value).
from dipy.direction import peaks_from_model
peak_indices = peaks_from_model(
model=dti_model, data=dwi_data, sphere=sphere, relative_peak_threshold=.2,
min_separation_angle=25, mask=dwi_mask, npeaks=2)
Additionally, we will apply a stopping criterion for our tracking based on the FA map. That is, we will stop our tracking when we reach a voxel where FA is below 0.2.
from dipy.tracking.stopping_criterion import ThresholdStoppingCriterion
stopping_criterion = ThresholdStoppingCriterion(fa_img, .2)
We will also need to specify where to “seed” (begin) the fiber tracking. Generally, the seeds chosen will depend on the pathways one is interested in modelling. In this example, we will create a seed mask from the FA map thresholding above our stopping criterion.
from dipy.tracking import utils
seed_mask = fa_img.copy()
seed_mask[seed_mask >= 0.2] = 1
seed_mask[seed_mask < 0.2] = 0
seeds = utils.seeds_from_mask(seed_mask, affine=affine, density=1)
Now, we can apply the tracking algorithm!
As mentioned previously, EuDX is the fiber tracking algorithm that we will be
using. The most important parameters to include are the indices representing
the discretized directions of the peaks (peak_indices), the stopping
criterion, the seeds, the affine transformation, and the step sizes to take
when tracking!
from dipy.tracking.local_tracking import LocalTracking
from dipy.tracking.streamline import Streamlines
# Initialize local tracking - computation happens in the next step.
streamlines_generator = LocalTracking(
peak_indices, stopping_criterion, seeds, affine=affine, step_size=.5)
# Generate streamlines object
streamlines = Streamlines(streamlines_generator)
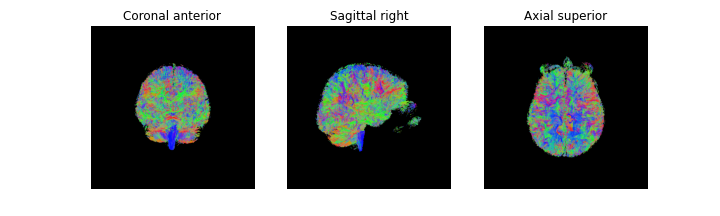
We just created a deterministic set of streamlines using the EuDX algorithm
mapping the human brain connectome (tractography). We can save the streamlines
as a Trackvis file so it can be loaded into other software for visualization
or further analysis. To do so, we need to save the tractogram state using
StatefulTractogram and save_tractogram to save the file. Note that we will
have to specify the space to save the tractogram in.
from dipy.io.stateful_tractogram import Space, StatefulTractogram
from dipy.io.streamline import save_tractogram
sft = StatefulTractogram(streamlines, dwi_img, Space.RASMM)
# Save the tractogram
save_tractogram(sft, os.path.join(out_dir, "tractogram_deterministic_EuDX.trk"))
We can then generate the streamlines 3D scene using the FURY python package,
and visualize the scene’s contents with Matplotlib.
from fury import actor, colormap
from utils.visualization_utils import generate_anatomical_volume_figure
# Plot the tractogram
# Build the representation of the data
streamlines_actor = actor.line(streamlines, colormap.line_colors(streamlines))
# Generate the figure
fig = generate_anatomical_volume_figure(streamlines_actor)
fig.savefig(os.path.join(out_dir, "tractogram_deterministic_EuDX.png"),
dpi=300, bbox_inches="tight")
plt.show()
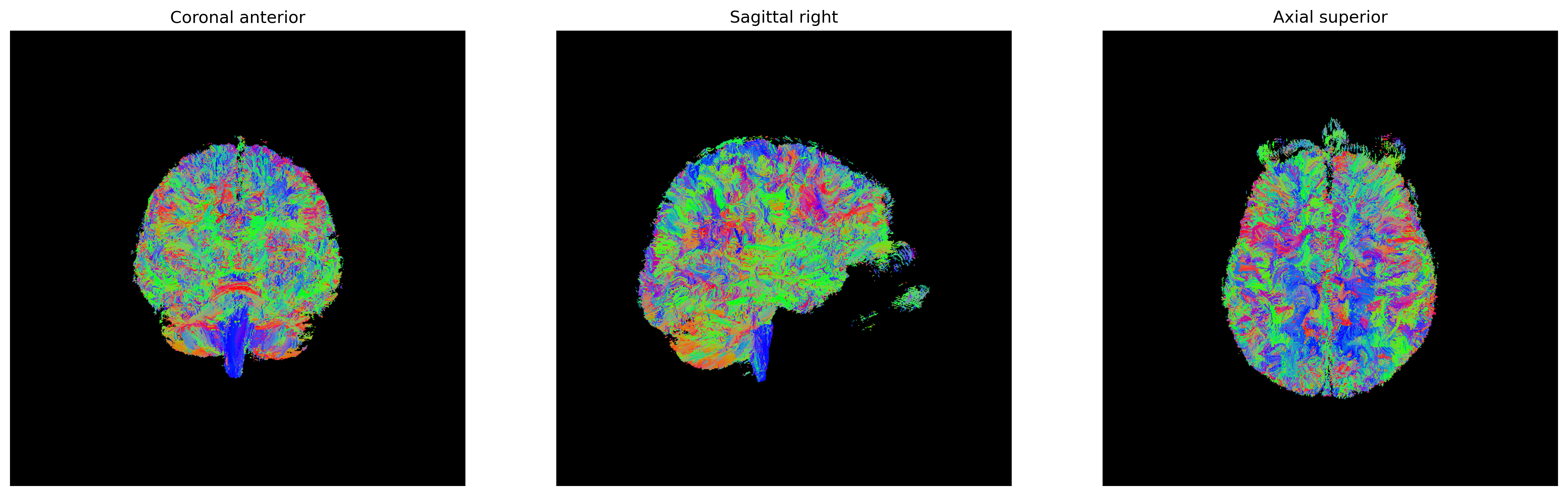
Exercise 1
In this episode, we applied a threshold stopping criteria to stop tracking when we reach a voxel where FA is below 0.2. There are also other stopping criteria available. We encourage you to read the
DIPYdocumentation about the others. For this exercise, repeat the tractography, but apply a binary stopping criteria (BinaryStoppingCriterion) using the seed mask. Visualize the tractogram!Solution
import os import nibabel as nib import numpy as np from bids.layout import BIDSLayout from dipy.io.gradients import read_bvals_bvecs from dipy.core.gradients import gradient_table from dipy.data import get_sphere from dipy.direction import peaks_from_model import dipy.reconst.dti as dti from dipy.segment.mask import median_otsu from dipy.tracking import utils from dipy.tracking.local_tracking import LocalTracking from dipy.tracking.streamline import Streamlines from utils.visualization_utils import generate_anatomical_volume_figure from fury import actor, colormap import matplotlib.pyplot as plt dwi_layout = BIDSLayout("../../data/ds000221/derivatives/uncorrected_topup_eddy", validate=False) gradient_layout = BIDSLayout("../../data/ds000221/", > > validate=False) # Get subject data subj = '010006' dwi_fname = dwi_layout.get(subject=subj, suffix='dwi', extension='.nii.gz', return_type='file')[0] bvec_fname = dwi_layout.get(subject=subj, extension='.eddy_rotated_bvecs', return_type='file')[0] bval_fname = gradient_layout.get(subject=subj, suffix='dwi', extension='.bval', return_type='file')[0] dwi_img = nib.load(dwi_fname) affine = dwi_img.affine bvals, bvecs = read_bvals_bvecs(bval_fname, bvec_fname) gtab = gradient_table(bvals, bvecs) dwi_data = dwi_img.get_fdata() dwi_data, dwi_mask = median_otsu(dwi_data, vol_idx=[0], numpass=1) # Specify the volume index to the b0 volumes # Fit tensor and compute FA map dti_model = dti.TensorModel(gtab) dti_fit = dti_model.fit(dwi_data, mask=dwi_mask) fa_img = dti_fit.fa evecs_img = dti_fit.evecs sphere = get_sphere('symmetric362') peak_indices = peaks_from_model( model=dti_model, data=dwi_data, sphere=sphere, relative_peak_threshold=.2, min_separation_angle=25, mask=dwi_mask, npeaks=2) # Create a binary seed mask seed_mask = fa_img.copy() seed_mask[seed_mask >= 0.2] = 1 seed_mask[seed_mask < 0.2] = 0 seeds = utils.seeds_from_mask(seed_mask, affine=affine, density=1) # Set stopping criteria stopping_criterion = BinaryStoppingCriterion(seed_mask==1) # Perform tracking streamlines_generator = LocalTracking( peak_indices, stopping_criterion, seeds, affine=affine, step_size=.5) streamlines = Streamlines(streamlines_generator) # Plot the tractogram # Build the representation of the data streamlines_actor = actor.line(streamlines, colormap.line_colors(streamlines)) # Generate the figure fig = generate_anatomical_volume_figure(streamlines_actor) plt.show()
Exercise 2
As an additional challenge, set the color of the streamlines to display the values of the FA map and change the opacity to
0.05. You may need to transform the streamlines from world coordinates to the subject’s native space usingtransform_streamlinesfromdipy.tracking.streamline.Solution
import numpy as np from fury import actor from dipy.tracking.streamline import transform_streamlines from utils.visualizations_utils import generate_anatomical_volume_figure import matplotlib.pyplot as plt streamlines_native = transform_streamlines(streamlines, np.linalg.inv(affine)) streamlines_actor = actor.line(streamlines_native, fa_img, opacity=0.05) fig = generate_anatomical_volume_figure(streamlines_actor) plt.show()
Key Points
Deterministic tractography methods perform tracking in a predictable way